Alternative splicing (AS) is a fundamental mechanism that regulates gene expression by generating multiple distinct mRNA isoforms from a single gene. This process enables the synthesis of diverse proteins from a limited number of genes, contributing to the complexity and versatility of the proteome. Understanding and detecting alternative splicing events are critical for comprehending cellular function, disease mechanisms, and the development of therapeutic interventions. Bioinformatics software plays a pivotal role in the identification and analysis of alternative splicing events, providing researchers with the tools necessary to explore splicing variations across different conditions, tissues, and species.
Introduction to Alternative Splicing
Alternative splicing occurs when different combinations of exons and introns are joined together during the post-transcriptional modification of pre-mRNA. This process can result in a variety of mRNA transcripts, each encoding a different protein isoform. There are several types of alternative splicing events, including:
- Exon skipping: The most common type of AS, where an exon is excluded from the final transcript.
- Mutually exclusive exons: Where one of two exons is included in the transcript, but not both.
- Alternative 5' or 3' splice sites: Alterations in the splice site selection at the 5' or 3' ends of an exon, leading to alternative isoforms.
- Intron retention: Where an intron is retained in the final transcript, resulting in a longer mRNA isoform.
- Alternative promoters or polyadenylation sites: Changes in the site of transcription initiation or termination, producing different isoforms of mRNA.
These variations in splicing patterns can significantly affect protein function, cellular processes, and disease development. Aberrant splicing is associated with many diseases, including cancer, neurodegenerative disorders, and genetic diseases.
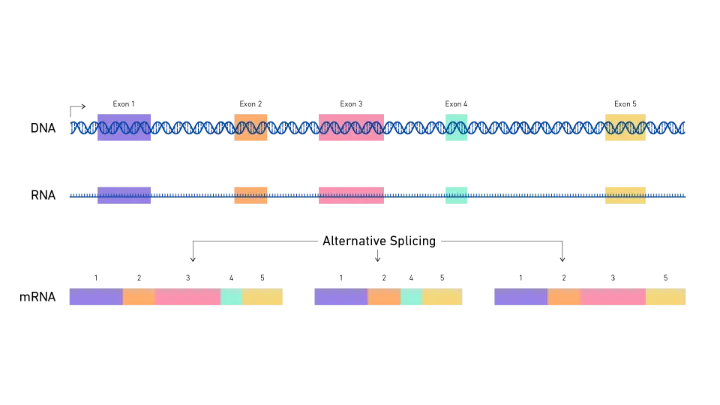
The Role of Bioinformatics in Alternative Splicing Detection
Bioinformatics tools are essential for detecting, quantifying, and analyzing alternative splicing events from RNA sequencing (RNA-Seq) data. RNA-Seq allows for the comprehensive and unbiased examination of transcriptomes, making it a powerful technique for studying alternative splicing. The vast amount of data generated by RNA-Seq, however, necessitates the use of specialized computational tools to identify splicing events accurately.
Bioinformatics software helps researchers in the following ways:
- Identifying Splice Junctions: By analyzing RNA-Seq reads, bioinformatics software can map read alignments to reference genomes and identify splice junctions that occur between exons and introns.
- Quantifying Splicing Events: Tools can quantify the abundance of different splice variants by measuring the expression levels of individual splice junctions or exons.
- Visualizing Splicing Patterns: Many tools provide graphical representations of splicing events, allowing researchers to easily interpret complex data.
- Comparing Splicing Across Conditions: Software can compare alternative splicing patterns across different conditions, tissues, or species, helping to identify splicing events that are biologically significant or disease-related.
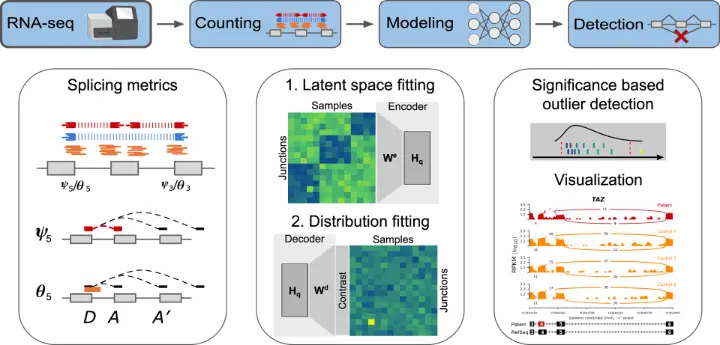
Popular Bioinformatics Tools for Alternative Splicing Detection
A variety of bioinformatics tools and software platforms are available for the detection and analysis of alternative splicing events from RNA-Seq data. These tools range from basic splice site prediction software to comprehensive platforms that provide detailed splicing analysis. Below are some of the most widely used tools:
1. TopHat and Cufflinks
- TopHat: A widely used tool for RNA-Seq alignment and splicing detection. It identifies splice junctions by aligning reads to the genome and detecting splice sites.
- Cufflinks: This tool works alongside TopHat and provides a comprehensive analysis of alternative splicing. It assembles the RNA-Seq reads into transcripts, quantifies transcript expression levels, and identifies splicing events.
These tools are often used together, with TopHat handling the alignment and splicing detection, and Cufflinks providing transcript assembly and quantification.
2. STAR (Spliced Transcripts Alignment to a Reference)
- STAR: An ultrafast RNA-Seq read aligner that handles spliced alignments efficiently. STAR can detect splice junctions directly from the RNA-Seq data and provides high accuracy for spliced read alignment. It is highly effective for large datasets, making it one of the most popular tools for RNA-Seq analysis.
- Integration with other tools: STAR can be paired with other software like DESeq2 or EdgeR for differential splicing analysis.
3. rMATS (Replicate Multivariate Analysis of Transcript Splicing)
- rMATS: A specialized tool designed for detecting differential alternative splicing events from RNA-Seq data. rMATS focuses on comparing splicing between different conditions and provides robust statistical analysis to determine significant splicing events.
- Key Features: rMATS is highly recommended for studies that involve comparing multiple conditions, such as cancer versus normal tissues, or different time points in developmental biology.
4. MAJIQ (Modeling Alternative Junctions in Quantitative Splicing)
- MAJIQ: A tool that models alternative splicing by reconstructing the entire set of isoforms present in a given RNA-Seq dataset. It is designed to handle complex splicing events and allows for the analysis of splicing at both the level of individual genes and across the entire transcriptome.
- Visualization: MAJIQ provides extensive visualization options, making it easier for researchers to interpret alternative splicing events.
5. SpliceSeq
- SpliceSeq: A tool designed specifically for visualizing and analyzing alternative splicing from RNA-Seq data. It is capable of identifying both known and novel splice junctions and providing insights into differential splicing across different conditions.
- User-Friendly Interface: SpliceSeq includes a web-based platform that allows users to perform splicing analysis without extensive computational expertise.
6. ASpli
- ASpli: A tool designed for the analysis of alternative splicing in a comparative context. It uses a reference genome to align RNA-Seq data and identify splicing events. ASpli is particularly effective for comparative splicing studies, where the goal is to identify differences in splicing between different conditions, tissues, or species.
7. DEXSeq
- DEXSeq: A differential exon usage analysis tool that allows researchers to examine the differential usage of exons across different conditions. DEXSeq is particularly useful for identifying splicing events that affect the inclusion or exclusion of specific exons in a transcript.
- Integration with other tools: DEXSeq can be used alongside other RNA-Seq analysis tools like DESeq2 to perform a comprehensive differential expression and splicing analysis.
8. SashimiPlot
- SashimiPlot: A tool designed to create visualizations of splicing patterns. It generates graphical representations of splice junctions, allowing researchers to see which exons are included or excluded in a given condition.
- Interactive Plots: SashimiPlot provides interactive plots that help researchers explore splicing patterns at the genomic level.
Data Integration and Interpretation
Bioinformatics tools for alternative splicing detection often output large volumes of data, which can be challenging to interpret without the proper context. Therefore, integrating splicing data with other types of omics data—such as gene expression, proteomics, or epigenomics—can provide more comprehensive insights into the biological significance of alternative splicing events.
For instance, combining splicing data with gene expression data can help identify genes whose splicing patterns change in response to external stimuli or disease states. Additionally, coupling splicing analysis with proteomics can offer insights into how alternative splicing leads to the production of distinct protein isoforms, which may have different functional properties.
Moreover, integrating splicing data with clinical data could open up new avenues for understanding how splicing variations contribute to disease progression and treatment responses. This could lead to the development of personalized therapies targeting specific splicing events, making alternative splicing an exciting area of research for future drug development.
Conclusion
Alternative splicing is a crucial regulatory mechanism that contributes to the diversity of the transcriptome and proteome. Bioinformatics software for alternative splicing detection has revolutionized our ability to study these events in-depth and has provided the tools necessary to understand their roles in development, disease, and cellular function. From splice junction identification to differential splicing analysis and visualization, a wide array of tools is available to researchers looking to explore the complexities of splicing.
As RNA-Seq technology continues to evolve and generate even larger datasets, the need for efficient, accurate, and scalable bioinformatics tools will only increase. By leveraging these tools, researchers can gain deeper insights into the regulatory mechanisms underlying gene expression, ultimately advancing our understanding of human biology and disease. With continued advancements in both bioinformatics and RNA-Seq technology, the future of alternative splicing detection holds great promise for unraveling the complexities of gene regulation and its implications for health and disease.